Help
*Best experienced using the Chrome or Safari browsers
1. Overview
2. Browse
3. Search
4. Visualize
5. Download
6. Submit
7. Contact Us
1. Overview
The singleCellBase is a high-quality manually curated database of cell markers for single cell annotation across multiple species developed by CapitalBiotech Technology (Figure 1). All the records origin from 2,247 high-quality publications using 10x Genomics, and will be updated periodically. We firstly manually surveyed all the full texts and relevant supplement tables, and then extracted the associations of marker genes and cell types which were observed and refined in the studies, as well as the descriptions of evidences. The first manually curation was followed by double-checking to ensure the accuracy and credibility. Finally, we provides a user-friendly interface to the scientific community to browse, search and download records of marker genes and cell types.
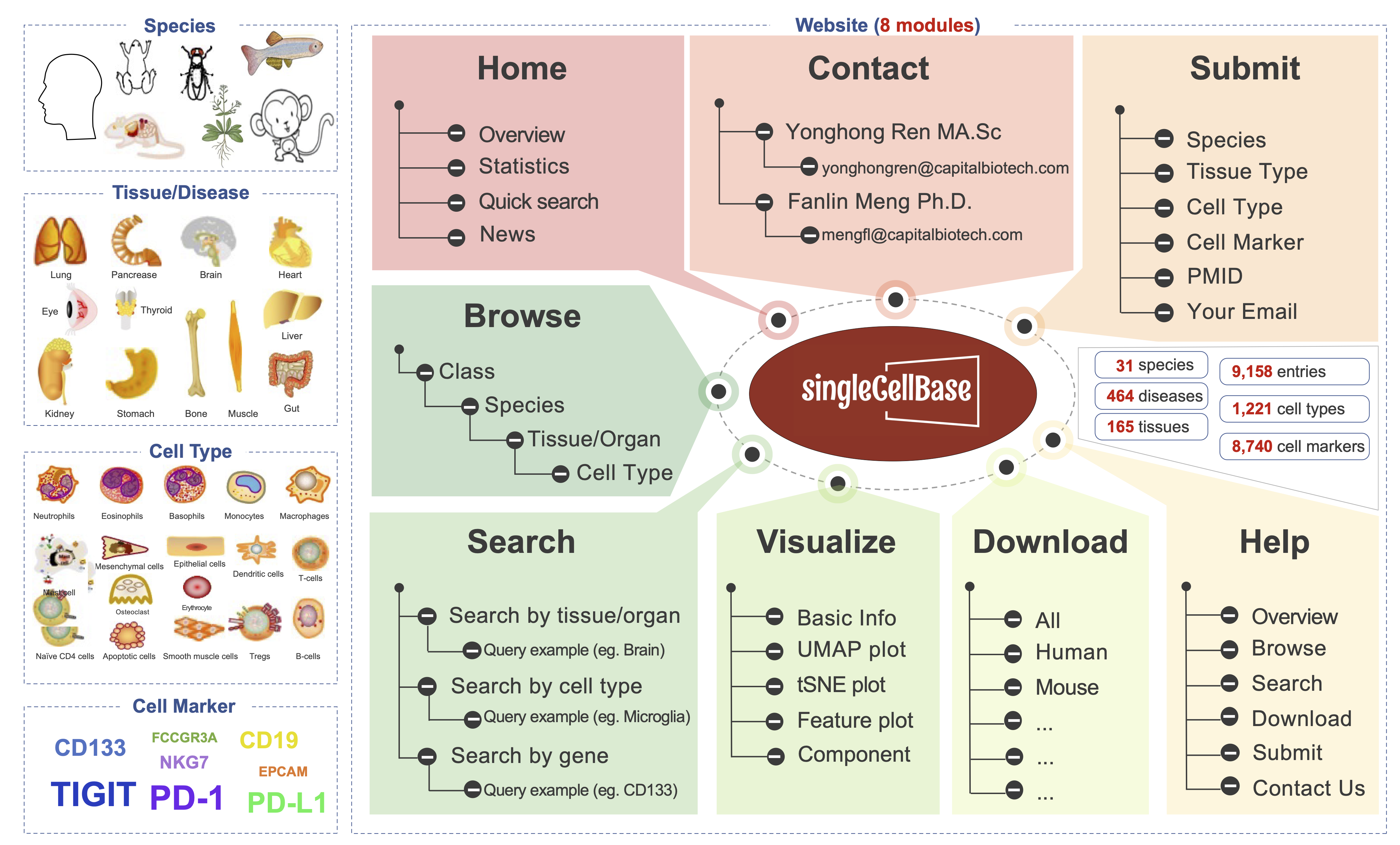
Figure 1. The overview of the singleCellBase database.
The singleCellBase is the first resource with marker genes for manual annotation across multiple species. The resource providing ineluctable prior knowledge required by manual cell annotation, which is valuable to interpret scRNA-seq data and elucidate what cell type or cell state that a cell population represents.
2. Browse
To browse cell types and marker genes association in the singleCellBase, please click the "Browse" component in the website navigation. All the records were divided into a three-level hierarchy structure according to species, tissues and cell types. Users can browse all entries by cell types. Taking a browse "B cells" in "Bone marrow" of "Human" as an example. To browse the entries, please subsequently select "Human", "Bone marrow ", and "B cells" option in the hierarchy structure. All the records relevant to B cells in Human bone marrow from multiple literature resources will be illustrated in the right panel as shown in Figure 2 left. The fields of "Tissue", "Cell Type" and "Cell Marker" could be the most important for users. The "PMID" field provide the link to the supporting literature in PubMed in each record. By clicking "More" in the "Details" field at the end of each record, the details of the supporting literature like "Journal", "SCI IF", "Evidence", "Title", and "GEO ID" will be shown in Figure 2 right.
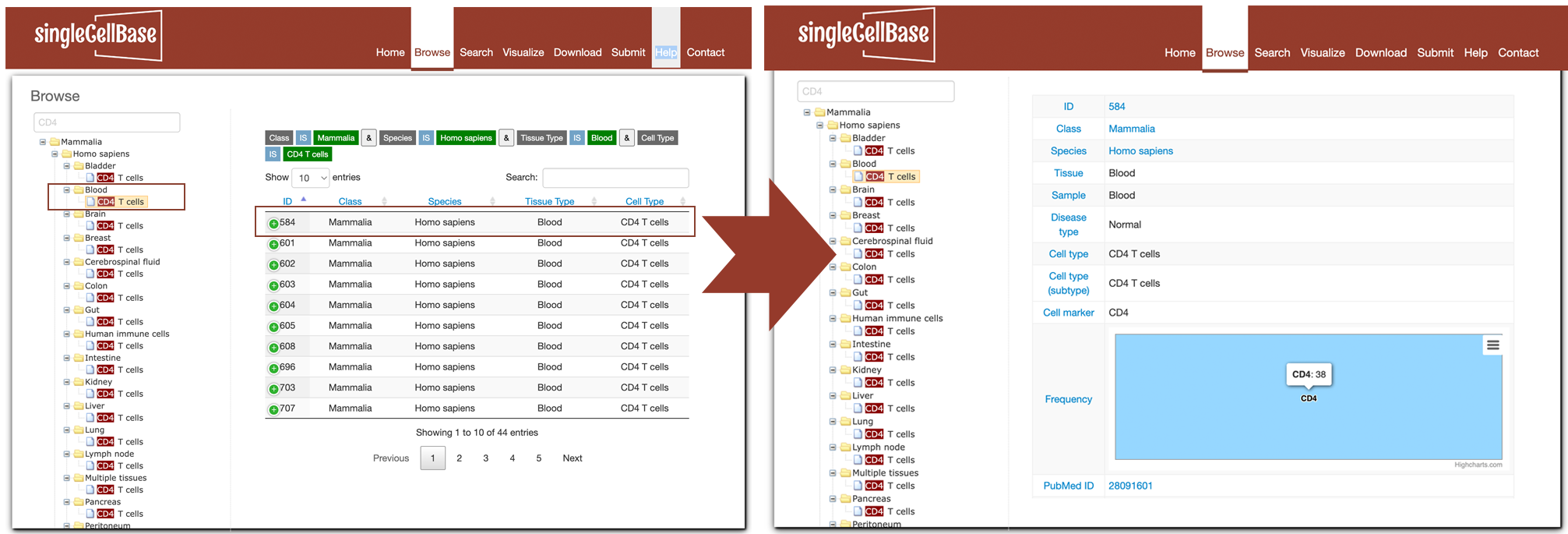
Figure 2. The 'Browse' interface of the singleCellBase database.
3. Search
To search records in the database, please click the "Search" component in the website navigation. 1. Users can search all records in three forms of key words: search by tissues, or cell types or cell markers (i.e. genes marked by symbols). After input any form of a key word, please click "submit" button to launch the search. Taking a search of "Brain" tissue as an example (Figure 3A). All the records relevant to Brain tissue across multiple species and cell types will be displayed in the result page as shown in Figure 3B. 2. It is noted that singleCellBase database also provides a fuzzy search for convenience. Taking a search of "Micro" as an example (Figure 3 left). All the records relevant to "Micro" across multiple species and tissues will be displayed in the result page as shown in Figure 3 right.
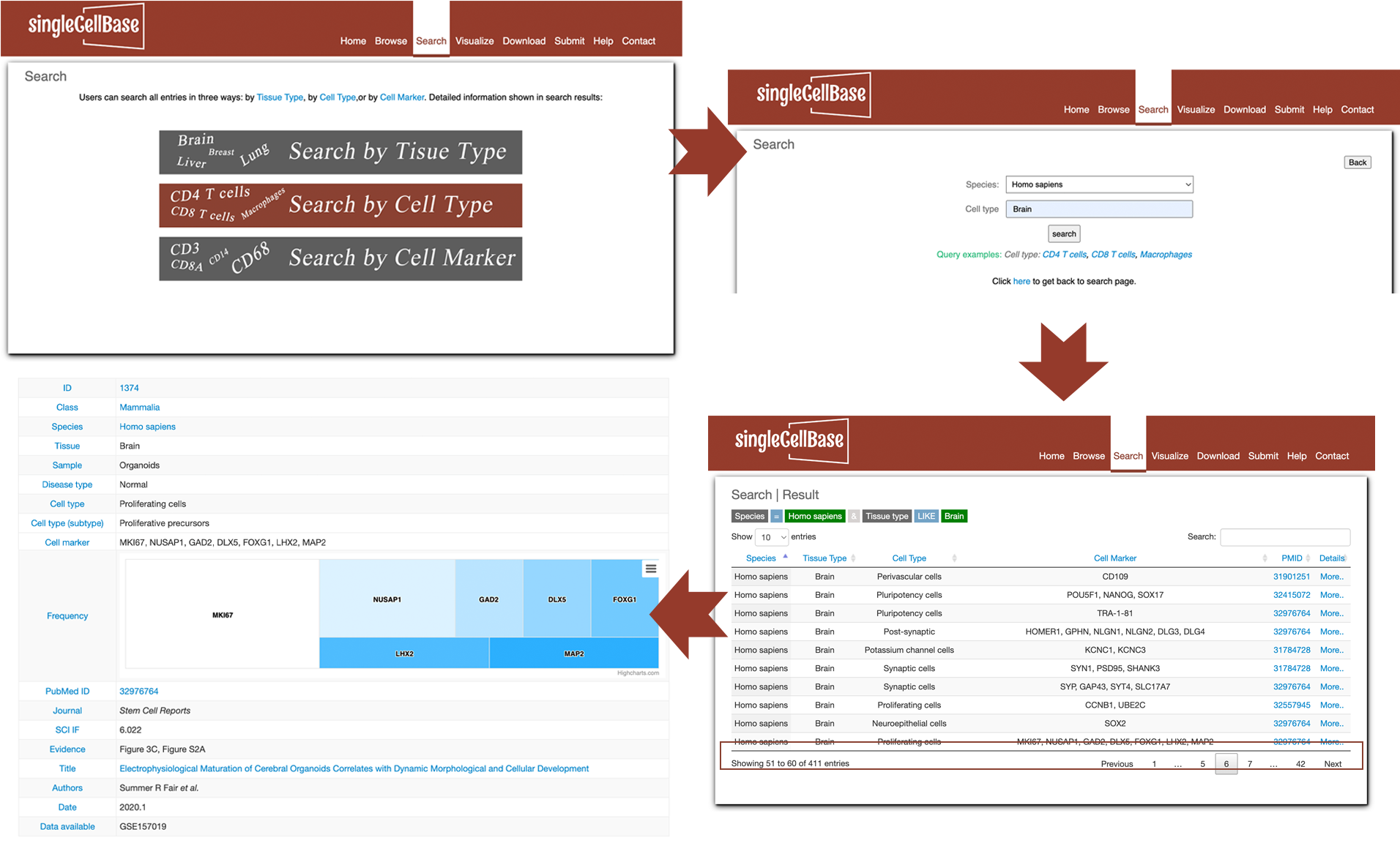
Figure 3. A illustration of the 'Search' component in the singleCellBase.
4. Visualize
We developed 'Visualize' module to visualize single-cell RNA-seq data. Please click the "Visualize" component in the website navigation. After select a dataset and input a gene, please click "submit" button to launch the search. Taking a "gse140628_pancreatic_cancer" dataset and a "EPCAM" gene as an example (Figure 4A). Basic information about dataset and several figures (e.g. UMAP plot, tSNE plot, Feature plot, cell component) will be displayed in the result page as shown in Figure 4B.
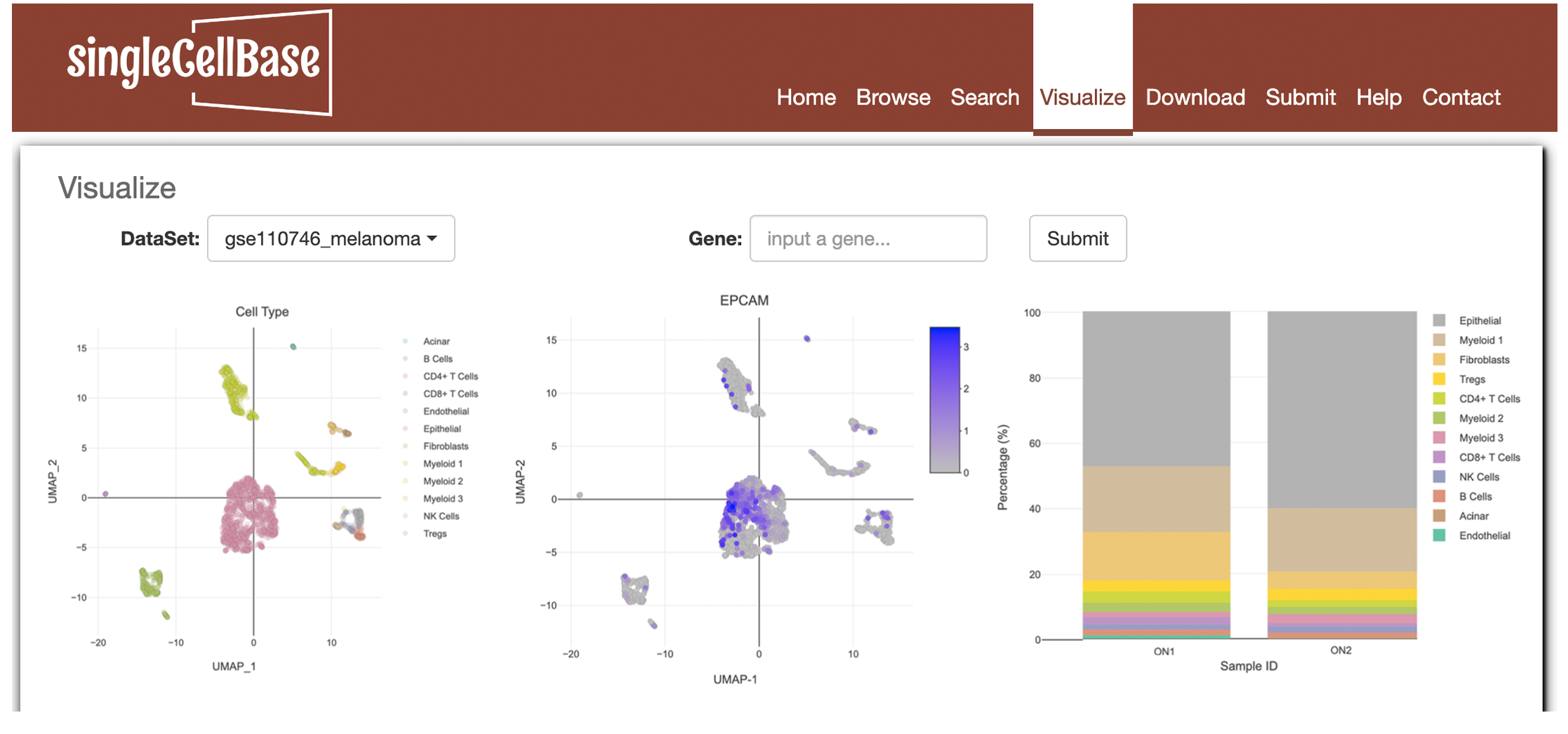
Figure 4. A illustration of the 'Visualize' component in the singleCellBase.
5. Download
To download data in the database, select the menu 'Download'. SingleCellBase database provides downloadable file in TXT format. Users can download all cell marker data sets, which include four files: 1) all cell markers of different cell types from different tissues in human and mouse, 2) cell markers in human, 3) cell markers in mouse, and 4) cell markers from single cell sequencing researches.
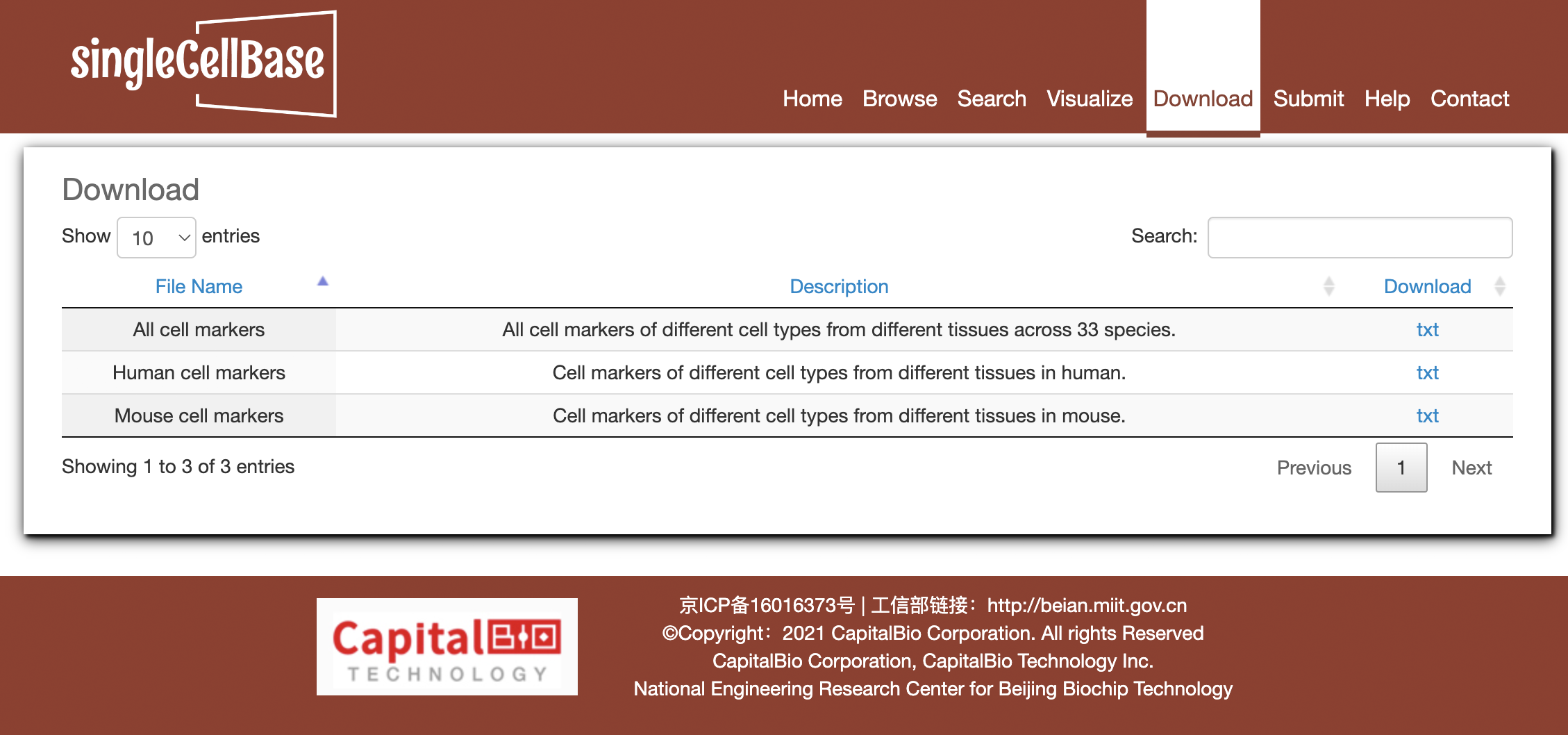
Figure 5. The 'Download' component in the singleCellBase.
6. Submit
If you have putative records that source from other resources or your prior knowledge and still not be harbored in our singleCellBase, please submit the valuable records into our database. The submitted records are required to contain fields of "Species", "Tissue", "Cell Type", "Cell Marker", "PMID" at least as shown in Figure 4.
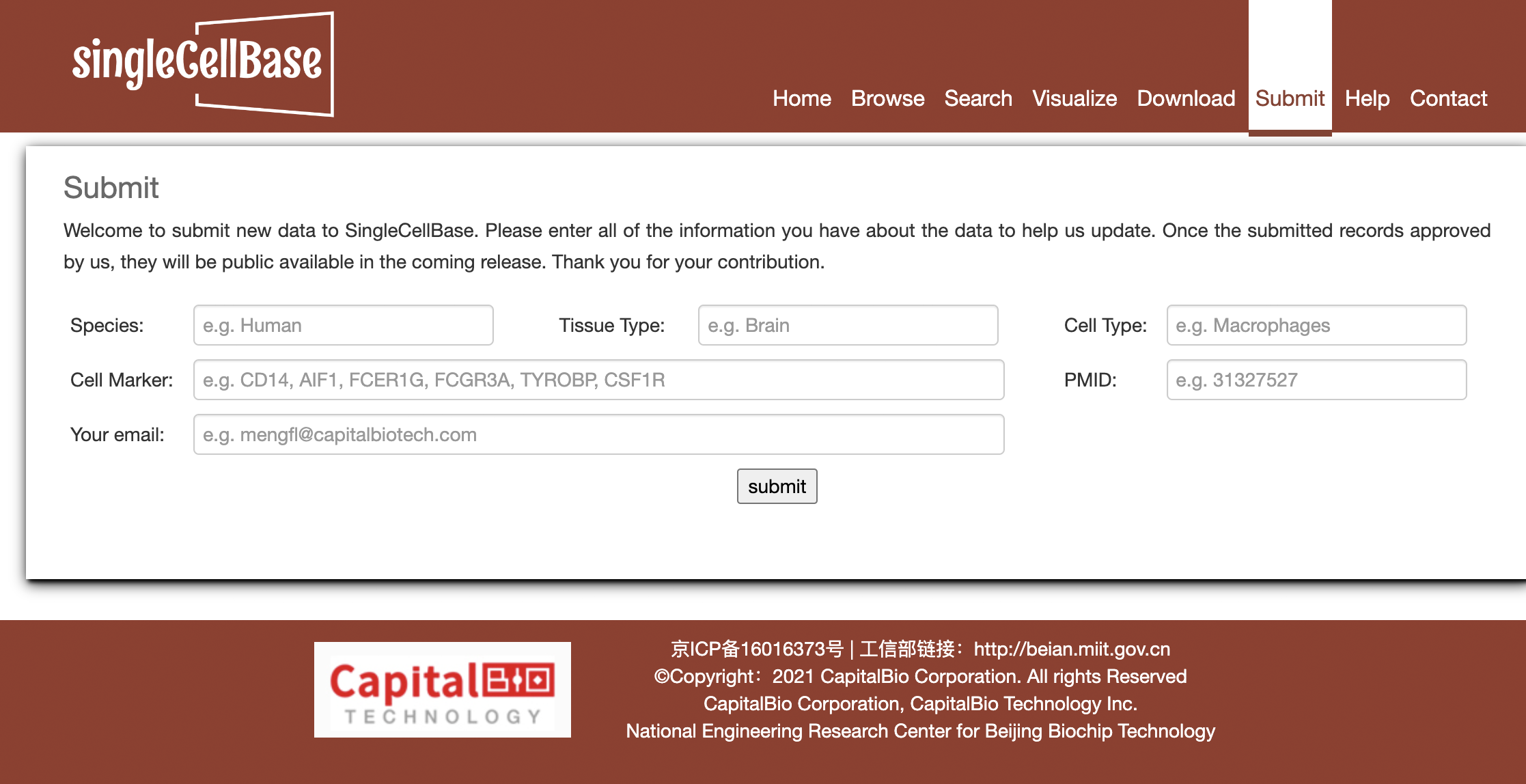
Figure 6. The 'Submit' component in the singleCellBase.
7. Contact Us
If you have any questions or comments regarding to SingleCellBase, you may contact us by sending an email to Yonghong Ren (yonghongren@capitalbiotech.com) or Fanlin Meng (mengfl@capitalbiotech.com).
